FAS2rDNA: From DNA annotation to FASTA-formatted sequence
Supports local (CLI version) and cloud (Colab version) implementations.
FAS2rDNA is publicly available
FAS2rDNA is publicly available on GitHub and the instructions for use are completely documented.
About FAS2rDNA
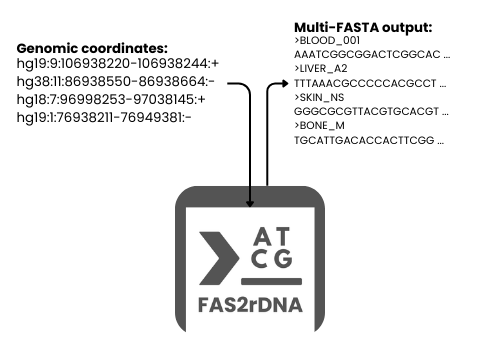
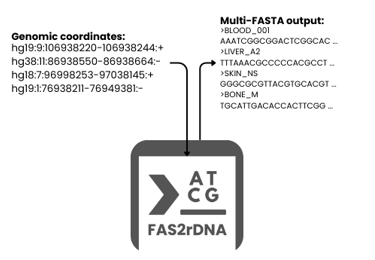
FAS2rDNA is a publicly-available tool for automated, strand-aware reconstruction of DNA sequences from genomic coordinates across multiple reference assemblies, supporting multi-species processing. It is designed for batch, multi-region, and mega-scale workflows in genomics, bioinformatics, and machine learning-driven sequence analysis.
What FAS2rDNA Does
FAS2rDNA simplifies the extraction and reconstruction of DNA sequences by transforming tabulated user data inputs into reconstructed multi-FASTA outputs. Given genomic coordinates, FAS2rDNA automatically retrieves reference genomes and reconstructs the corresponding DNA sequences in a reproducible and deterministic manner. The tool is optimized for large datasets where manual sequence extraction or ad hoc scripting becomes inefficient, error-prone, or non-reproducible.
Why FAS2rDNA Matters
Modern genomics is no longer limited by data generation, but by how efficiently genomic information can be transformed into machine-usable representations. As sequencing projects scale to millions of regions and thousands of samples, reproducible and automated sequence reconstruction becomes a critical bottleneck. FAS2rDNA addresses this gap by providing a deterministic, genome-scale bridge between coordinate-based genomic data and sequence-level analysis—enabling downstream applications in large-scale genomics, machine learning, and AI-driven biological modeling.
How to Use FAS2rDNA
FAS2rDNA works both in local and cloud-based implementations.
1. CLI version: Designed for high-performance and production-scale workflows.
Intended for local machines, servers, and HPC environments
Supports large batch jobs and automation
Ideal for pipelines, reproducible research, and integration with downstream tools
Learn more here: https://github.com/mahvin92/FAS2rDNA
2. Colab version: Designed for accessibility, teaching, and rapid prototyping.
Runs entirely in the browser
No local setup required
Ideal for demonstrations, tutorials, and exploratory analysis
Learn more here: https://github.com/mahvin92/FAS2rDNA-Colab
Publications
FAS2rDNA-Colab: A cloud-based workflow for pan-cancer, isoform-wide miRNome reconstitution across TCGA cohorts. Protocols.io (2025). DOI: 10.17504/protocols.io.14egn1xr6v5d/v1
High-throughput isoform-wide miRNome sequence reconstruction in the TCGA-LUAD cohort using FAS2rDNA. Protocols.io. (2025). DOI: 10.17504/protocols.io.rm7vzenqxvx1/v1





